
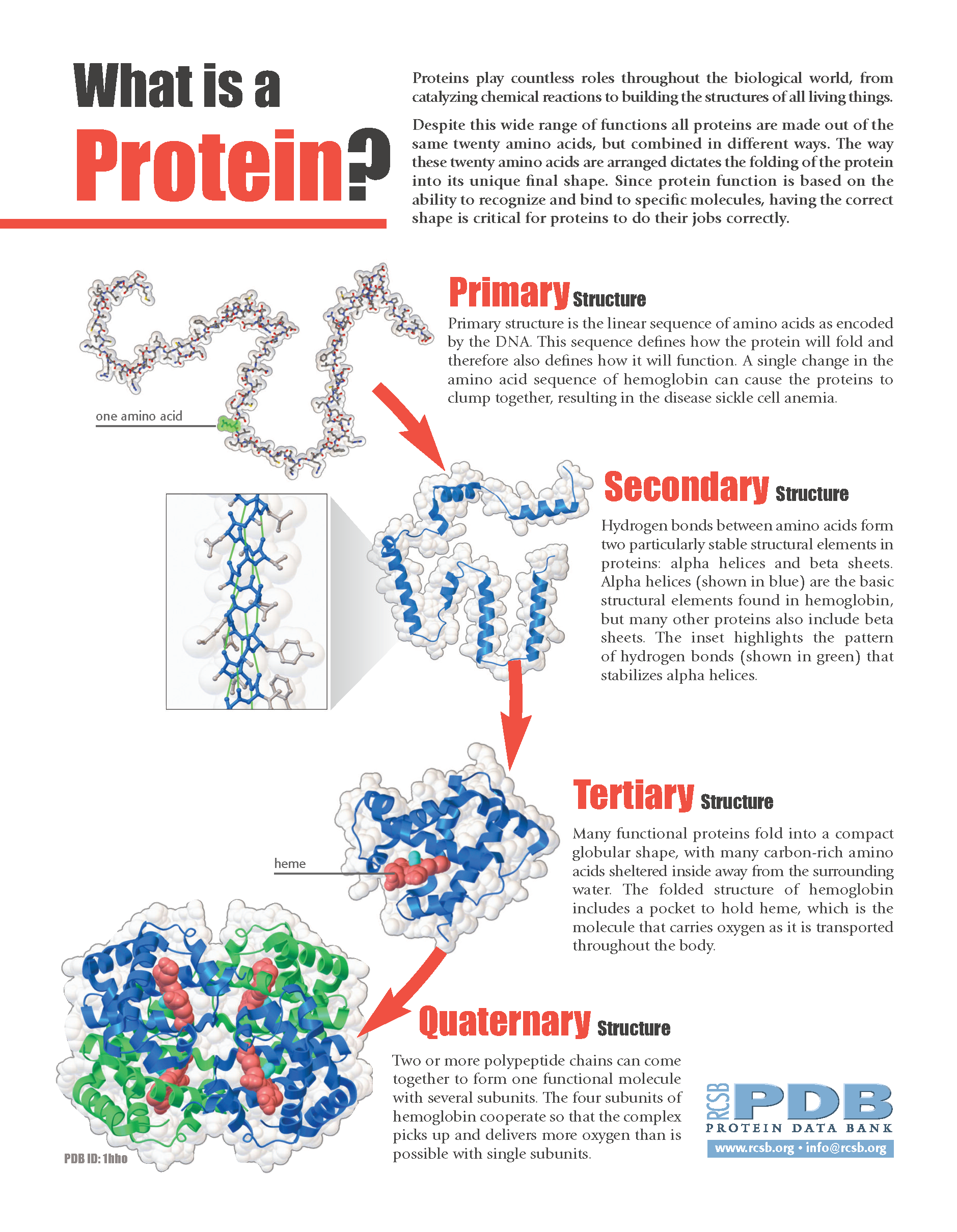

The protein secondary structure is the bridge of three-dimensional structures and sequences, which is determined by the effect of hydrogen bonds in the polypeptide chain ( Rafid et al., 2020 Grmez et al., 2021 Guo et al., 2021 Sharma and Srivastava, 2021 Singh et al., 2021). Hence, it is important for computer scientists to be able to predict the three-dimensional structures of proteins from their sequences rapidly and relatively inexpensively (Uniprot Yang et al., 2016). The three-dimensional structure of a protein can be obtained by X-ray crystallography, multi-dimensional magnetic resonance, and cryo-electron microscopy, which are expensive and time-consuming, and these data are generally provided in the Protein Data Bank (PDB) ( PDB, 1971 Berman and Henrick Nakamura, 2003 Kim et al., 2008). The function of a protein depends on its three-dimensional structure, which is determined by the protein sequence and folding activities within a living cell ( Zou, 2000). Proteins play important roles in life activities, such as signal transduction and transmission, living material transportation, catalysis, and immunity ( Saini and Hou, 2013 Pka et al., 2021). The experimental results indicate that the conditional GAN-based protein secondary structure prediction (CGAN-PSSP) model is workable and worthy of further study because of the strong feature-learning ability of adversarial learning. Then, we propose a PSSP method based on the proposed multiscale convolution module and ICA module. We introduce a new multiscale convolution module and an improved channel attention (ICA) module into the generator to generate the secondary structure, and then a discriminator is designed to conflict with the generator to learn the complicated features of proteins.
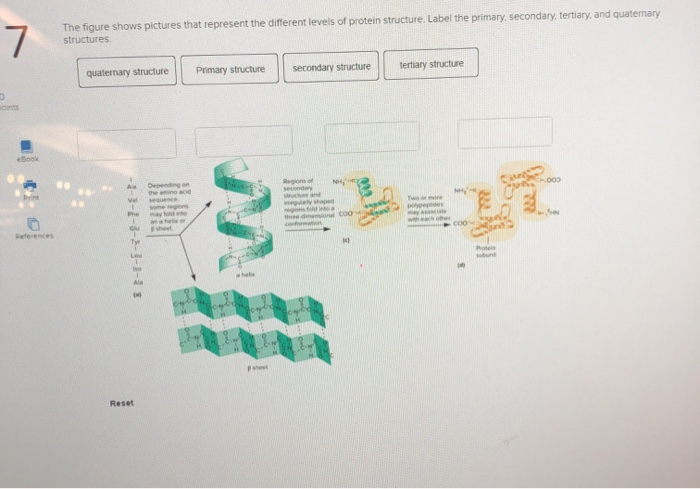
To explore a new technique of PSSP, this study introduces the concept of an adversarial game into the prediction of the secondary structure, and a conditional generative adversarial network (GAN)-based prediction model is proposed. Driven by deep learning, the prediction accuracy of the protein secondary structure has been greatly improved in recent years. Protein secondary structure prediction (PSSP) aims to construct a function that can map the amino acid sequence into the secondary structure so that the protein secondary structure can be obtained according to the amino acid sequence. Prediction of the protein secondary structure is a key issue in protein science.


 0 kommentar(er)
0 kommentar(er)
